Challenge Description
This challenge aims to encourage participants to develop novel approaches that can effectively handle negative statements in knowledge graphs (KGs).
Since ontologies are already able to express negation and the enrichment of biomedical KGs with interesting negative statements is gaining traction, this challenge focuses on exploring ontology-rich biomedical KGs. These KGs use an ontology to provide rich descriptions of real-world entities instead of focusing on describing relations between entities themselves. Furthermore, there is an essential difference between a positive and a negative statement related to the implied inheritance in this kind of KG. A positive statement between an entity and an ontology class implies a positive statement between that entity and all the superclasses of the ontology class. Conversely, a negative statement between an entity and an ontology class implies a negative statement between the entity and all the subclasses of the ontology class.
Participants in this challenge will be evaluated on three relation prediction tasks. Relation prediction is the task of learning a relation between two KG entities (a pair) when the relation itself is not explicitly defined in the KG.
References:
Important Dates
March 16, 2024: Call for challenge
March 30, 2024: Release of KGs and datasets
July 28, 2024: Submission of systems
August 5, 2024: Submission of reports
August 19, 2024: Notification of Acceptance
November 11-15, 2024: Challenge presentation during ISWC
Tasks Description
The challenge encompasses three distinct biomedical relation prediction tasks: Protein-Protein Interaction Prediction, Gene-Disease Association Prediction, and Disease Prediction. For each task, the goal is to predict whether a relation exists given two biomedical KG entities (which can belong to the same or different KGs). The relations between the entities are not explicitly defined in the KG. Therefore, this is a fundamentally distinct task from link prediction, where the training set relations are part of the KG.
To tackle these relation prediction tasks, common approaches typically employ three steps: (1) generate embeddings for each entity in the KG; (2) aggregate the embeddings of each entity in a pair into a single representation; (3) use these aggregated representations as input to a binary classifier to learn a relation prediction model.
For each task, participants will be provided with the KG in OWL format, a set of entity pairs with information regarding the existence of a relation (training dataset), as well as a list of entity pairs where information about the existence of a relation is not given (test dataset).
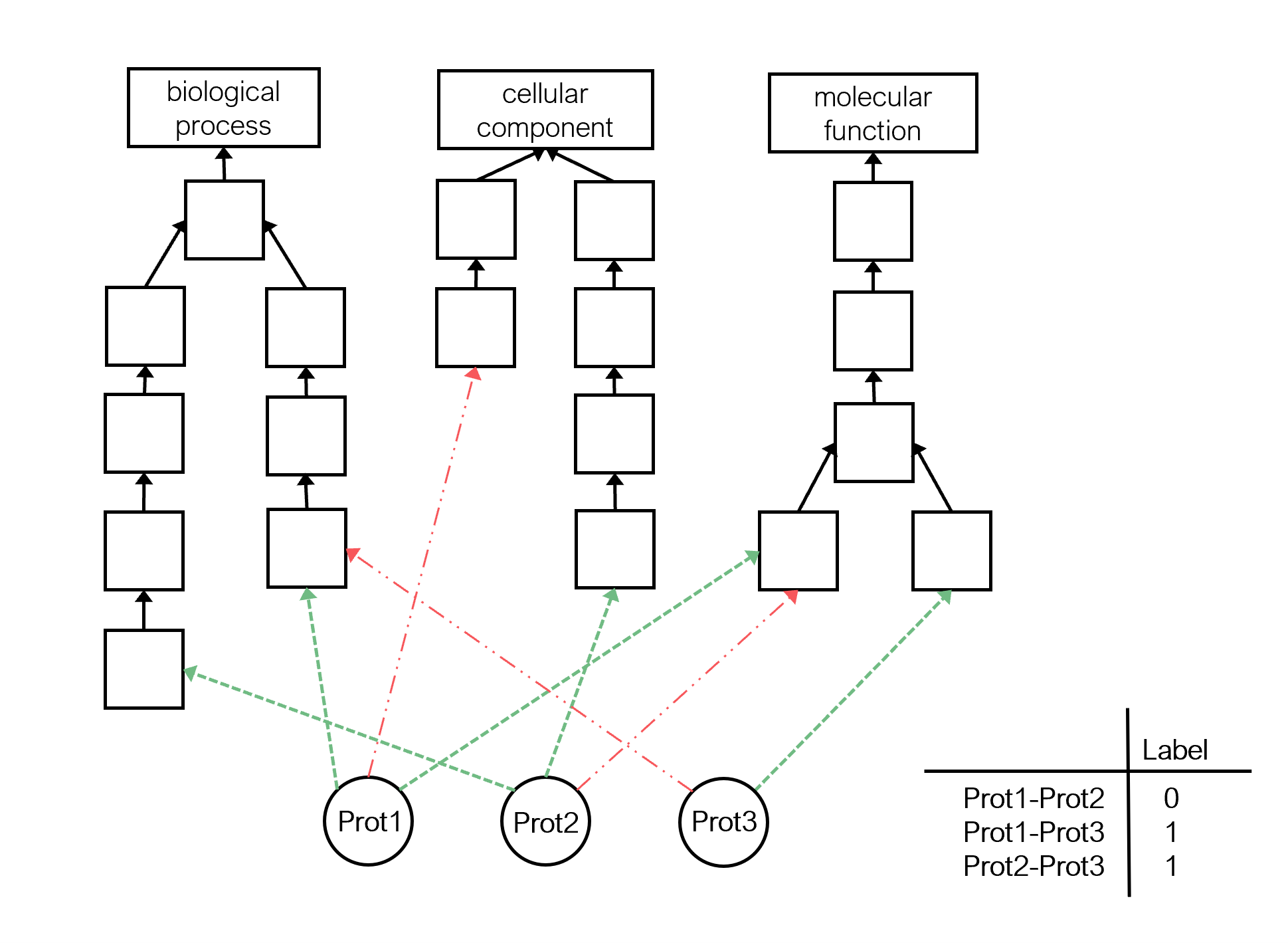
Predicting if there is an interaction between two proteins is a fundamental task in molecular biology for understanding biological systems.
The proteins are described in the Gene Ontology (GO) KG. This KG is built by integrating three sources:
- The GO that contains more than 50000 ontology classes that describe gene products according to the molecular functions they perform, the biological processes they are involved in, and the cellular components where they act.
- The positive statements between proteins and GO classes.
- The negative statements between proteins and GO classes.
The target protein pairs are extracted from the STRING database.
| Training set | Test set | |
|---|---|---|
| Instances | 440 | 209 |
| Positive pairs | 1024 | - |
| Negative pairs | 1024 | - |
| Positive statements | 7364 | 3952 |
| Negative statements | 8476 | 3912 |
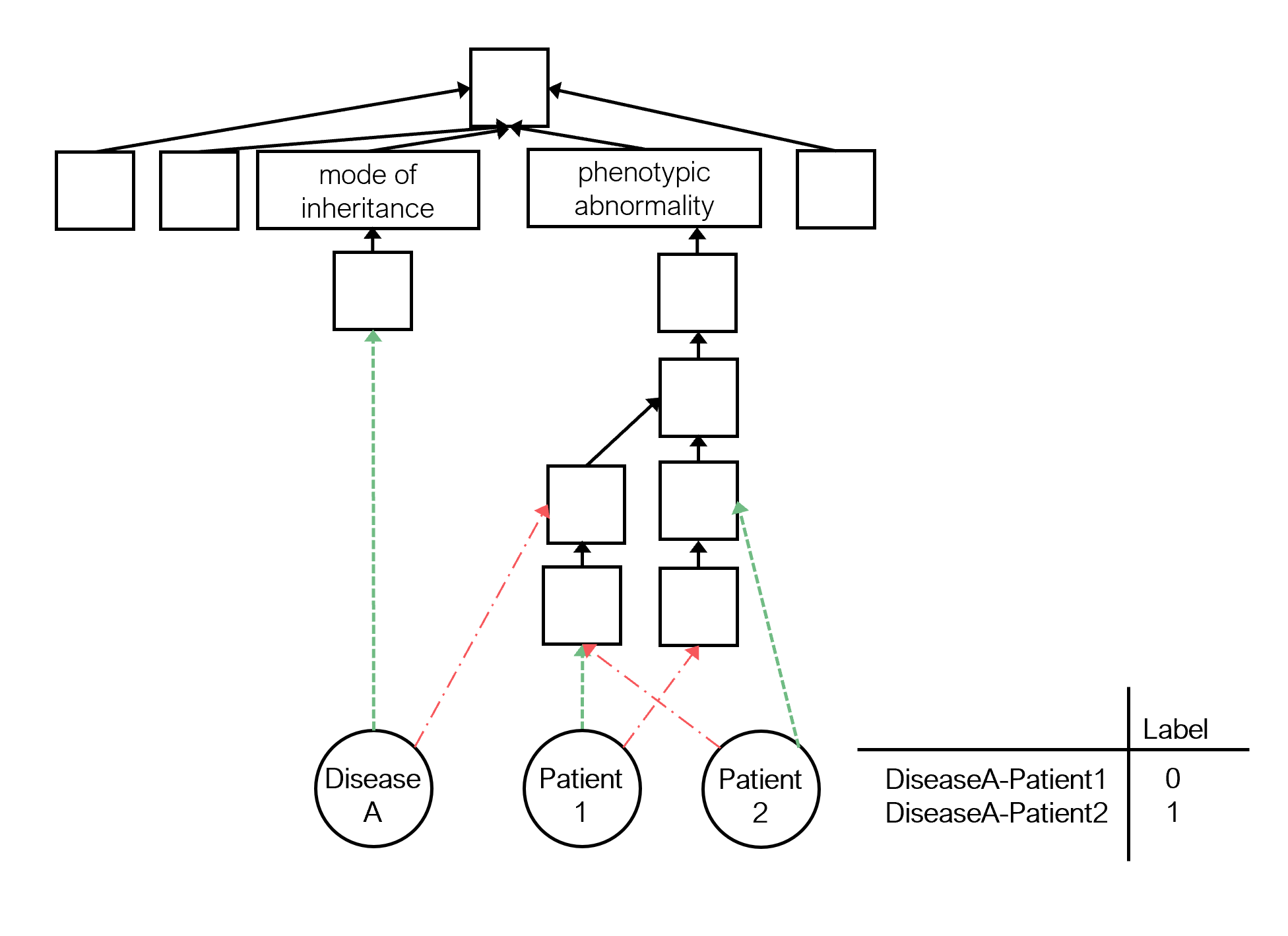
Predicting if a synthetic patient has been diagnosed with a specific disease is an essential and complicated task that must be executed accurately and efficiently.
The patients and diseases are described in the Human Phenotype Ontology (HP) KG. This KG is built by integrating:
- The HP characterizes phenotypic abnormalities discovered in human hereditary diseases according to five semantic aspects: phenotypic abnormalities, mode of inheritance, clinical course, clinical modifier and frequency.
- The positive and negative statements between patients and HP classes.
- The positive and negative statements between diseases and HP classes.
The target patient-disease pairs are generated using a synthetic data methodology.
| Training set | Test set | |
|---|---|---|
| Instances | 1693 | 1330 |
| Positive pairs | 660 | - |
| Negative pairs | 681120 | - |
| Positive statements | 40823 | 32977 |
| Negative statements | 223 | 151 |
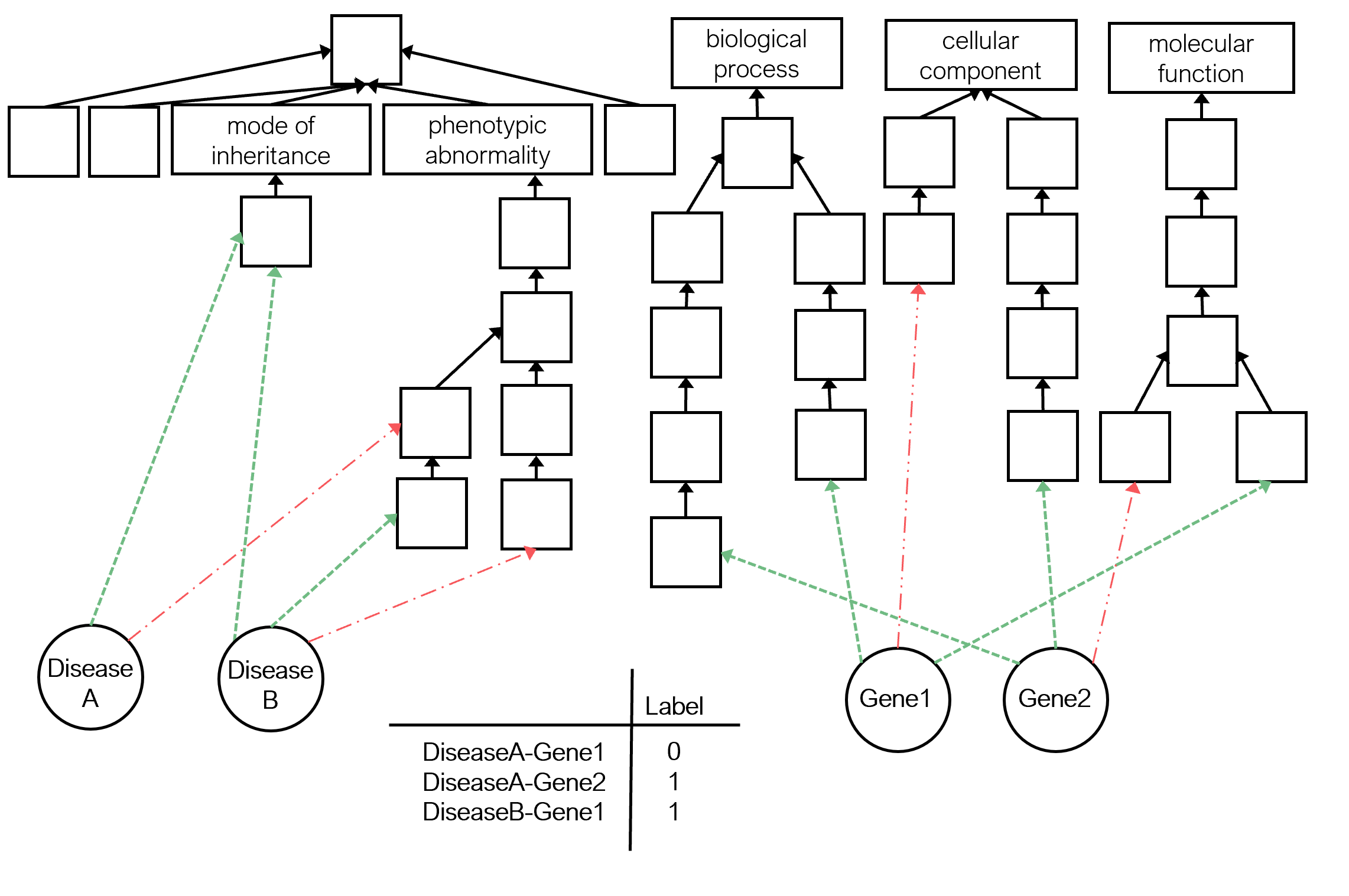
Predicting if there is an association between a gene and a disease is crucial to understanding the disease mechanisms and recognising potential biomarkers or therapeutic targets.
Two KGs are used to describe the pair entities. Genes are described under the Gene Ontology (GO) KG, and diseases are described under the Human Phenotype Ontology (HP) KG.
- The GO KG contains GO, the positive and negative statements between genes and GO classes.
- The HP KG contains HP, the positive and negative statements between diseases and HP classes.
The target relations to predict are extracted from DisGeNET database.
| Training set | Test set | |
|---|---|---|
| Instances | 281 | 78 |
| Positive pairs | 107 | - |
| Negative pairs | 107 | - |
| Positive statements | 5663 | 1546 |
| Negative statements | 2179 | 392 |
Submission details
The submissions should be sent to challengenegknow@gmail.com and should include the group name and a list of full names, positions, affiliations, and mails of all the participants in the group.
Participants are required to submit:
- A CSV file with the predicted labels (0 or 1) for each test instance. Please use the template file as a reference for the correct format of the prediction file.
- A report detailing the approach they employed. The report should be in PDF format (4-10 pages, CEUR Workshop style).
The participants are also strongly encouraged to submit their code implementations to facilitate reproducibility.
Evaluation
For each task, the different submissions will be evaluated with respect to the prediction performance (recall, precision and weighted average F-measure) in the test set.
Organizing Commitee

Cátia Pesquita
LASIGE, Faculdade de Ciências da Universidade de Lisboa
Heiko Paulheim
Data and Web Science Group, Universität Mannheim
Rita T. Sousa
Data and Web Science Group, Universität MannheimContacts
Email:
challengenegknow@gmail.com